
Sanger DNA Sequencing

Our DNA Sequencing division functions both as a small-scale sequencing facility and a center for large-scale sequencing projects. It offers more than 18 years of solid experience in conventional Sanger DNA sequencing, including specialty sequencing and specific supporting services, and reliably provides high-quality services with rapid turnaround and competitive pricing.
Overview
-
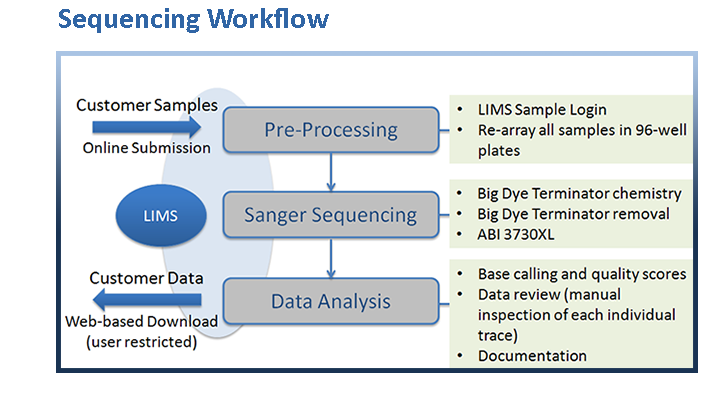
Our Sanger sequencing process begins with a cycle sequencing reaction using the Applied Biosystems BigDye v3.1 Cycle Sequencing Kit which employs a fluorescently-labeled dideoxy-nucleotide chain termination method to generate extension products from DNA templates. Extension products are purified using SPRI technology. Subsequently, fragment separation and sequence detection is carried out by capillary electrophoresis on the 96-well capillary matrix of an ABI3730XL DNA Analyzer, followed by post-detection processing. In the final analysis step, a combination of software base calling and manual inspection of the individual trace files is employed to warrant the highest possible quality of the generated data.
-
Upon successful completion of the analysis, automated email notifications are sent to our customers who are then able to access their sequencing data securely through our website by logging into our LIMS and downloading the results (chromatograms, text files, and quality files are returned to the customer).
-
Our in-house Laboratory Information Management System (LIMS) tracks each individual sample (or ‘order’) from sample entry and reception through the entire sequencing pipeline, and it allows our customers to submit orders online, check the status of their order(s),and finally to retrieve their data.
-
To ensure consistency in quality during all stages of our process, the core has implemented Standard Operating Procedures (SOPs), stringent multi-level quality controls (in-process quality controls, routinely performed out-of-process QC exercises, controlled reagent release into production), thorough Failure-Mode-Effect-Analysis (FMEA), daily tracking of quality metrics, and continuous employee training programs for all stages of the process.
-
In the rare event of an equipment failure, of a reagent failure, or of a human error, all affected samples will be automatically repeated at no extra cost for our customers.
-
We provide extensive customer support on an individual basis as well as technical and scientific consultation expertise with the goal of improving the success rate of our customers’ research experiments. Our support includes but is not limited to assistance in template and sample preparation, in the selection of the appropriate sequencing protocol, and in data interpretation and troubleshooting if needed.
-
The core makes sequencing a convenient daily service for the Mass General Brigham community by providing free local sample-pickup (with the exception of weekends and official MGH Holidays). With our daily courier service between our facility, the MGH main and Charlestown campuses, and the Simches building, samples are delivered within an hour, processed immediately, and data returned within 24 hours (business days only).
-
The sequencing group is capable of processing up to 2500 samples per day. To accelerate larger sequencing projects (typically between 1,000-20,000 samples), operational flexibility combined with rapid changes in production scheduling allows for an even higher daily throughput.
Once sequenced, customer samples will be stored for 1 week to enable re-ordering of complete plates or single tubes.
Sequencing Services
Individual Tube Sequencing
This service option accommodates project sizes as small as a single sample. The customer will submit a premix of DNA template (purified plasmids or purified PCR product) and sequencing primer, prepared in a tube as described in our Sample Submission section. Dependent on the template, read lengths of up to 750 bp can be usually achieved.
96-well Plate (High-Throughput) Sequencing
This service employs the same state-of-the-art sequencing technology as used in our single-tube sequencing Service. Premixed samples (purified plasmids or purified PCR products) are submitted in plate-format (95 samples per plate). Well H12 remains empty to allow for the addition of an in-process control.
In support of any type of high-throughput DNA sequencing, the core offers 96-Well Plasmid DNA Preparation as well as 96-Well PCR Product Purification as add-on services (please refer to section Supporting Sanger Services for more details). In these cases, the primer(s) will be submitted separately (in tube or plate format) and added to the purified and normalized template(s) by the core personnel.
Due to the decreased processing time of samples provided in 96-well format, the core is able to offer a bulk-rate discount. For large-volume orders (> 10 96-well plates), we usually recommend a pilot experiment (1-2 plates typically) to determine optimal template and primer concentration and to select the most appropriate sequencing protocol.
Please contact the Core Director for more information and/or to discuss your project(s).
Difficult Template Sequencing
In Sanger sequencing, some templates/regions are difficult to sequence when using the standard dye-chemistry techniques and thus often fail to produce satisfactory reads. These templates include GC-rich regions (>60-65% GC content), repeats (di- and tri-nucleotide stretches), hairpin structures (shRNA), long homopolymer stretches (Poly A/T regions), and regions with strong stop sites. While our standard sequencing protocol may perform satisfactorily (but not necessarily reproducibly) for borderline cases, we are recommending our alternative sequencing protocol which can solve many of these problems and increase the read length for a wide range of these types of difficult templates. This service can be added on to both our standard individual tube and our 96-well plate sequencing services for an additional fee.
Long Read Sanger Sequencing
Recently, our sequencing unit implemented a Long-Read Sanger Sequencing service as part of its newly launched Custom Molecular Biology & Sequencing Service portfolio. Compared to our standard sequencing protocol (typical Q20 read length > 700 bases, dependent on template), this service delivers an additional 200 bases on average and can yield up to 1000 Q20 bases per sample. A minimum of 12 samples has to be submitted to take advantage of this service. Turnaround is between 24 and 48 hours. Please note that the rates for the new long-read service (details here) differ from those of our standard sequencing fees and reflect slightly increased costs due to the implementation of a separate workflow, increased instrument time, and processing of partially filled plates.
Capital Equipment
Our core facility is equipped with the latest state-of-the-art instrumentation to ensure both high consistency and high quality throughout our line of sequencing services
- ABI3730XL 96-capillary DNA Analyzer (3)
- Tecan Freedom EVO 200 Liquid Handling Robot (1)
- Eppendorf Mastercycler pro (20)
